Published online Jul 27, 2022. doi: 10.4254/wjh.v14.i7.1438
Peer-review started: November 9, 2021
First decision: January 9, 2022
Revised: January 24, 2022
Accepted: July 6, 2022
Article in press: July 6, 2022
Published online: July 27, 2022
Processing time: 260 Days and 3.4 Hours
Dermatologic adverse events (DAEs) are associated with a better outcome in patients with hepatocellular carcinoma (HCC) irrespective of the therapeutic agent received. The exact mechanisms associated with the development of DAEs are unknown although several studies point to direct toxicity of tyrosine kinase inhibitors (TKIs) to the skin or an immune-mediated reaction triggered by the oncologic treatment. As is the case in other conditions, individual genetic variants may partially explain a higher risk of DAEs.
To evaluate the contribution of several gene variants to the risk of developing DAEs in HCC patients treated with TKIs.
We first analyzed 27 single-nucleotide polymorphisms (SNPs) from 12 genes selected as potential predictors of adverse event (AE) development in HCC patients treated with sorafenib [Barcelona Clinic Liver Cancer 1 (BCLC1) cohort]. Three additional cohorts were analyzed for AGT1 (rs699) and AGT2 (rs4762) polymorphisms-initially identified as predictors of DAEs: BCLC2 (n = 79), Northern Italy (n = 221) and Naples (n = 69) cohorts, respectively. The relation between SNPs and DAEs and death were assessed by univariate and multivariate Cox regression models, and presented with hazard ratios and their 95% confidence intervals (95%CI).
The BCLC1 cohort showed that patients with arterial hypertension (AHT) (HR = 1.61; P value = 0.007) and/or AGT SNPs had an increased risk of DAEs. Thereafter, AGT2 (rs4762) AA genotype was found to be linked to a statistically significant increased probability of DAEs (HR = 5.97; P value = 0.0201, AA vs GG) in the Northern Italy cohort by multivariate analysis adjusted for BCLC stage, ECOG-PS, diabetes and AHT. The value of this genetic marker was externally validated in the cohort combining the BCLC1, BCLC2 and Naples cohorts [HR = 3.12 (95%CI: 1.2-8.14), P value = 0.0199, AGT2 (rs4762) AA vs AG genotype and HR = 2.73 (95%CI: 1.18-6.32) P value = 0.0188, AGT2 (rs4762) AA vs GG genotype]. None of the other gene variants tested were found to be associated with the risk of DAE development.
DAE development in HCC patients receiving TKIs could be explained by the AGT2 (rs4762) gene variant. If validated in other anti-oncogenic treatments, it might be considered a good prognosis marker.
Core Tip: Dermatologic adverse events (DAEs) are associated with a better outcome in patients with hepatocellular carcinoma (HCC) irrespective of the therapeutic agent received. Our study shows that DAE development in these patients can be explained by individual genetic variants in the AGT2 gene. AGT2 (rs4762) AA genotype was associated with DAE risk in the Northern Italy cohort and was externally validated in a cohort combining the BCLC1, BCLC2 and Naples cohorts. Therefore, DAE development in HCC patients receiving TKIs can be explained by the AGT2 (rs4762) gene variant. If validated in other anti-oncogenic treatments, it might be considered a good prognosis marker.
- Citation: Sapena V, Iavarone M, Boix L, Facchetti F, Guarino M, Sanduzzi Zamparelli M, Granito A, Samper E, Scartozzi M, Corominas J, Marisi G, Díaz A, Casadei-Gardini A, Gramantieri L, Lampertico P, Morisco F, Torres F, Bruix J, Reig M. Polymorphism AGT2 (rs4762) is involved in the development of dermatologic events: Proof-of-concept in hepatocellular carcinoma patients treated with sorafenib. World J Hepatol 2022; 14(7): 1438-1458
- URL: https://www.wjgnet.com/1948-5182/full/v14/i7/1438.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i7.1438
Treatment-related dermatologic adverse events (DAEs) are reported in a great number of oncological therapies. The profile and timing of on-target skin adverse events (AEs) varies across treatments and cancer types. In this regard, hand-foot skin reaction (HFSR) reported in patients receiving tyrosine kinase inhibitor (TKI) therapy resembles the already described hand-foot syndrome (HFS) described in patients treated with cytotoxic chemotherapies[1,2]. Moreover, several studies have described the association between DAE development and better patient outcome, and this association has been reported for different therapies [TKI, monoclonal antibody directed against EGFR[3] or immunotherapy[4,5] and different cancer types such as colorectal, renal, prostate, non-small cell lung and breast cancer as well as melanoma and hepatocellular carcinoma (HCC)][6]. Therefore, it appears that the association between DAE development and better patient outcome is observed regardless of cancer type or oncological treatment.
Although there are several hypotheses explaining the potential mechanisms of DAE development, the exact mechanisms remain unknown. Previous studies postulated that direct toxicity of TKIs to the skin could depend on drug secretion into eccrine glands[7] somehow copying the already described detection of doxorubicin in the sweat of treated patients[8]. Apart from other speculative explanations, inhibition of proangiogenic pathways could potentially prevent vascular repair mechanisms from functioning correctly and causing HFSR in high pressure areas that may be repeatedly exposed to subclinical trauma[9]. This would be applicable mainly to anti-angiogenic treatments but would leave other therapies out. Considering other drug treatments, a study on immune checkpoint inhibitors (ICIs) therapy in non–small cell lung cancer patients suggested that T cells would recognize antigens shared by both lung tumors and skin[10]. Consequently, treatment would target both organs thus leading to tumor regression associated with autoimmune skin toxic effects. However, the low frequency of tumors harboring potent neoantigens clearly compromises the rationale of this hypothesis. More recently, a study published by Ruiz-Pinto and colleagues[11] described the association between CDH4 genetic variants with the risk of developing capecitabine-induced HFS. In that study, CDH4 gene downregulation negatively impacted skin barrier function.
In 2018, we demonstrated that 91.6% of HCC patients who received sorafenib and achieved complete radiological response also developed DAEs within the first 2 mo of treatment[12,13]. Recently published data obtained in our group allowed us to identify the potential role of TKI in peripheral immune cell population profile modification towards a more pro-inflammatory behavior and phenotype[14]. Thus, we envision skin toxicity as a consequence of an immune-mediated reaction triggered by the oncologic treatment in patients prone to developing this side effect.
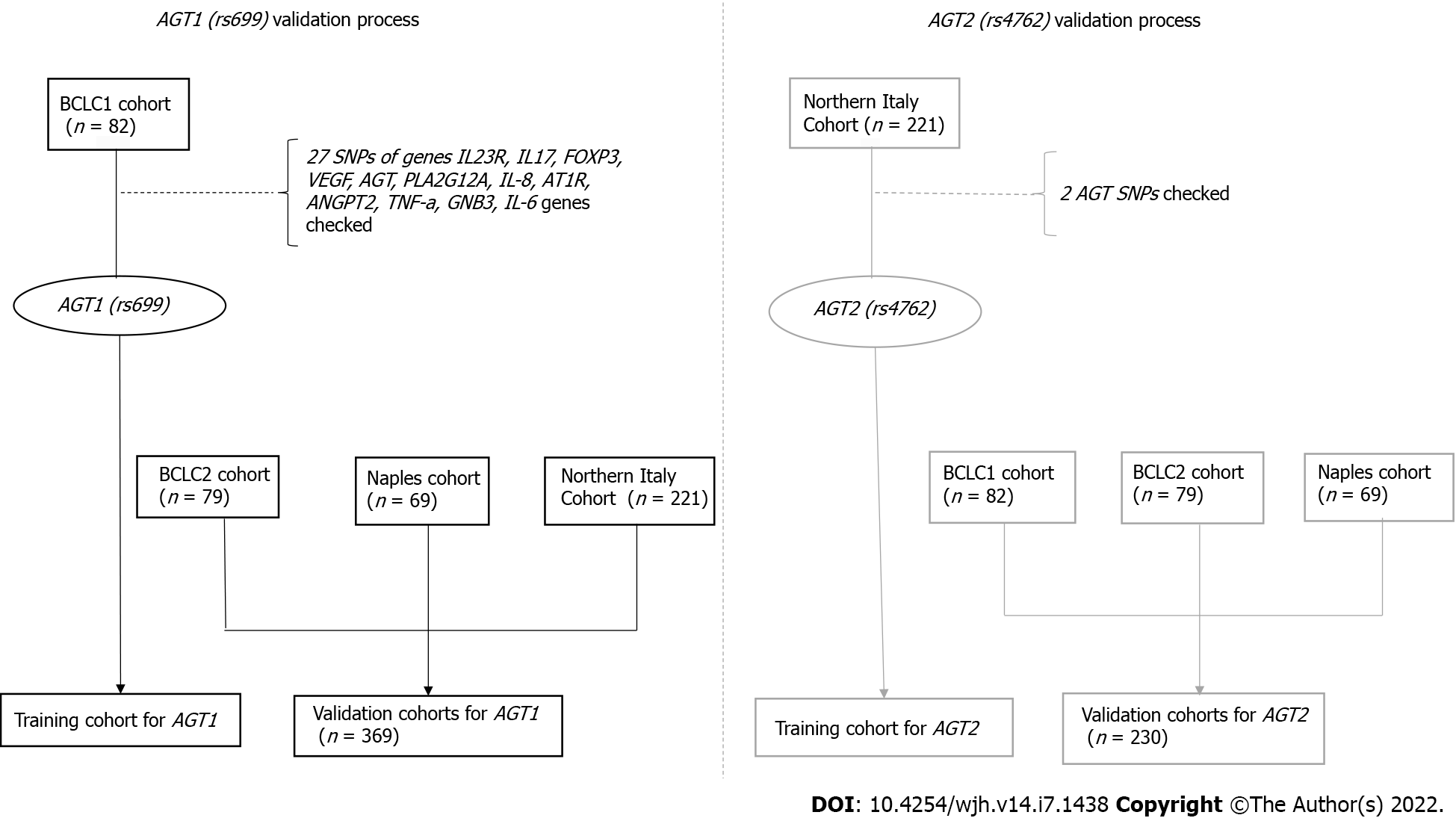
In order to uncover potential mechanisms underlying individual genetic susceptibility to AEs with clinical implications for risk prediction, we first analyzed 27 Single-Nucleotide Polymorphisms (SNPs) in 12 different genes as potential predictors of AE development in a Barcelona Clinic Liver Cancer 1 (BCLC1) cohort of 82 HCC patients treated with sorafenib. Upon identification of the potential relevance of the angiotensin genes, which include AGT1 (rs699) and AGT2 (rs4762), as predictors of DAEs, we further explored the association in three additional cohorts: a second BCLC cohort (n = 79), a Northern Italy cohort (n = 221) and a Naples cohort (n = 69).
Four cohorts of patients were analyzed in this study, two prospective cohorts from BCLC1 and BCLC2, and two additional cohorts from Northern Italy [Milan, Bologna, Meldola (FC) and Cagliari Hospitals] and Naples (Figure 1).
The study was approved by the institutional review board of each center (HCB/2009/4755, HCB/2015/0352, Ethical Board 2 480_2018 and CE/2014/193) and complied with the provisions of the Good Clinical Practice guidelines and the Declaration of Helsinki. A Data Transfer Protocol (DTP) was written according to the European regulation [General Data Protection Regulation (GDPR) 2016/679] and approved by each cohort responsible.
BCLC1 cohort: This cohort included patients referred to BCLC between February 2009 and March 2015 for sorafenib treatment.
Inclusion criteria were: (1) HCC diagnosed according to EASL guidelines[15]; (2) advanced HCC following the BCLC staging system or patients with earlier stages who could not benefit from treatments of higher priority; (3) normal liver or compensated cirrhosis with preserved liver function (Child-Pugh score ≤ 7 points without clinical ascites and/or encephalopathy; (4) performance status 0-1; (5) controlled arterial hypertension (AHT) and/or stable peripheral vascular disease; (6) adequate hematologic profile (platelet count > 60 × 109/L; hemoglobin > 8.5 g/dL; and prothrombin time > 50%); (7) adequate hepatic function (albumin > 2.8 g/dL; total bilirubin ≤ 3 mg/dL; and alanine and aspartate aminotransferases ≤ 5 times the upper limit of the normal range); and (8) adequate renal function (serum creatinine ≤ 1.5 times the upper limit of the normal range).
Exclusion criteria were: (1) Myocardial infarction in the last year or active ischemic heart disease; (2) acute variceal bleeding in the last month; (3) severe peripheral arterial disease; (4) arrhythmia under treatment with drugs different from beta-blockers or digoxin; (5) uncontrolled ascites; and (6) encephalopathy. All patients provided written informed consent before enrolment.
Follow-up: Clinical and laboratory assessments were performed monthly and radiologic tumor evaluation at week 4 and every 8 wk thereafter. Unscheduled visits due to AEs occurred according to patients’ needs.
DAEs were graded according to version 3.0 of the CTCAE of the National Cancer Institute, during treatment and 30 days after the last dose. We focused on DAEs within the first 60 days (eDAE) +/-7 days of treatment, which determined dose modification.
BCLC2 cohort: This cohort included patients referred to BCLC between June 2015 and August 2018 for sorafenib treatment.
The inclusion and exclusion criteria as well as the follow-up of this cohort were the same as for the BCLC1 cohort.
Northern Italy cohort: The Northern Italy cohort included patients with HCC treated with sorafenib prospectively enrolled between July 2008 and June 2018 in four tertiary centers in Italy whose data have already been published in several multicenter studies on sorafenib treatment[16,17]. Briefly, all patients with advanced HCC or intermediate-stage HCC refractory to or unsuitable for locoregional therapies, either histologically proven or diagnosed according to the AASLD guidelines (American Association for the Study of Liver Diseases 2005) and receiving sorafenib were eligible for analysis. Exclusion criteria were those established by the Italian Medicines Agency (AIFA), i.e., a performance status score > 2 and clinical decompensation. All patients received sorafenib with the standard schedule (400 mg bid continuously) with dose reduction applied as clinically indicated.
Follow-up: Follow-up consisted of a physical examination and complete blood count every 3 wk and Computed Tomography (CT) /Magnetic Resonance Imaging (MRI) scanning every 8 wk or as clinically indicated. Each visit included the recording of AEs, clinical laboratory tests, physical examination, and assessment of vital signs. At any time during treatment, the patient could have direct access to physicians for AE management. Safety was assessed in all patients who received at least one dose of sorafenib; AEs were graded according to the National Cancer Institute’s Common Terminology Criteria (version 3.0 CTCAE). Hepatic function deterioration was defined as a Child-Pugh score increase ≥ 2 points, which was evaluated at each visit and at predefined time points of week 12 and 24 of therapy. In line with the aim of the study, independently of clinical practice, we focused on the AEs which determined dose modification within the first 30 and 60 days of treatment, respectively. Treatment with sorafenib was continued until disease progression, unacceptable toxicity, or death. In each patient, the medical history, physical examination, blood cell count, serum chemistry, coagulation and alpha-fetoprotein levels were obtained at baseline and every 4 wk thereafter.
Naples cohort: This cohort included patients referred to the Gastroenterology Unit of the University Hospital Federico II of Naples between January 2014 and December 2019 for sorafenib treatment.
Inclusion criteria were: (1) HCC diagnosed according to EASL guidelines[15]; (2) advanced HCC following the BCLC staging system or patients with earlier stages who could not benefit from treatments of higher priority; (3) normal liver or compensated cirrhosis with preserved liver function (Child-Pugh score ≤ 7 points without clinical ascites and/or encephalopathy; (4) performance status 0-1; (5) controlled AHT and/or stable peripheral vascular disease; (6) adequate hematologic profile (platelet count > 30 × 103/L; hemoglobin > 8.5g/dL; and INR < 1.7; (7) adequate hepatic function (albumin > 2.8 g/dL; total bilirubin < 3 mg/dL; and alanine and aspartate aminotransferases < 5 times the upper limit of the normal range); and (8) adequate renal function (serum creatinine < 1.5 times the upper limit of the normal range).
Exclusion criteria were: (1) Myocardial infarction in the last year or active ischemic heart disease; (2) acute variceal bleeding in the last month; (3) severe peripheral arterial disease; (4) arrhythmia under treatment with drugs different from beta-blockers or digoxin; (5) uncontrolled ascites; and (6) encephalopathy. All patients provided written informed consent before enrolment.
Follow-up: Clinical and laboratory assessments were performed monthly and radiologic tumor evaluation at week 8 and every 8 wk thereafter. Unscheduled visits due to AEs occurred according to patients’ needs.
DAEs were graded according to version 3.0 of the CTCAE of the National Cancer Institute, during treatment and 30 days after the last dose. We focused on DAEs within the first 60 days (eDAE)+/-7 days of treatment, which determined dose modification.
Genomic DNA (gDNA) purification: gDNA was purified from isolated peripheral blood mononuclear cells (PBMCs) in the BCLC cohorts of patients and from 500 mL of whole frozen blood in the Naples cohort. gDNA purification was performed using the PureLink gDNA mini kit (Invitrogen, Thermo Fisher Scientific) following the manufacturer's instructions.
BCLC1 cohort: Patients were genotyped for a series of SNPs in IL23R, IL17, FOXP3, VEGF, AGT, PLA2G12A, IL-8, AT1R, ANGPT2, TNF-a, GNB3, and IL-6 genes. SNPs were selected according to reported associations with susceptibility to cardiovascular disease, hypertension, stroke, inflammatory pathways or even cancer development. The genes and SNPs analyzed are detailed in Supple
Twenty ng of gDNA were used for each SNP reaction. All SNPs were evaluated by means of TaqMan predesigned genotyping assays (Applied Biosystems, Thermo Fisher Scientific) and the procedure was performed following the manufacturer's instructions. A list of assays used is specified in Supple
Briefly, TaqMan® MGB probes from the genotyping assay provide a fluorescent signal for the amplification of each allele. SNP genotyping uses a 60 s extension time at 60ºC for 40 cycles. Real-time PCR software plots the results of the allelic discrimination data as a scatter plot of Allele 1 (VIC® dye) vs Allele 2 (FAM™ dye). Each well of the 96-well reaction plate is represented as an individual point on the allelic discrimination plot. Positive controls were used for each homozygote and heterozygote genotype.
Patients from the BCLC2, Northern Italy and Naples cohorts were genotyped for 2 SNPs of the AGT-gene [AGT1 (rs699) and AGT2 (rs4762)] using the TaqMan endpoint-genotyping assay, following the same techniques as previously described.
The statistical methods and analysis of this study were performed by Víctor Sapena and reviewed by Ferran Torres from the Hospital Clínic de Barcelona.
Quantitative variables are expressed as median and interquartile range [IQR 25th-75th percentiles]. Categorical variables are described as absolute frequencies and percentages (%).
Time to event variables are expressed as median and 95% confidence intervals (95%CI) using the Kaplan-Meier method. The log-rank test was used to compare Kaplan-Meier curves. Univariate and multivariate Cox regression models were used to estimate Hazard Ratios (HR) and 95%CI to evaluate the increased probability of developing grade II or early dermatologic events (eDAEs), dermatologic events (DAEs) or death according to each SNP. The multivariate adjusting factors were previously selected according to their clinical relevance, and these were BCLC stage (A or B vs C), ECOG-PS (0 vs ≥ 1), history of AHT (No vs Yes) and history of diabetes (No vs Yes). An analysis using 67 days as the landmark timepoint was used to calculate overall survival (OS) according to eDAE.
The level of significance was set at the two-tailed 5% level and all analyses and data base integration structure were performed with SAS 9.4 software (SAS Institute, Cary, NC, United States).
This study included 82 patients from the BCLC1 cohort, 79 from the second BCLC2 cohort, 221 from the Northern Italy cohort, and 69 from the Naples cohort.
Tables1, 2 and 3 describe the characteristics, OS and follow-up at the time of locking the database (December 2019) and the AE rates of all patients included in the study.
| BCLC1 cohort | BCLC2 cohort | Northern Italy cohort | Naples cohort | |
| Patients, n | 82 | 79 | 221 | 69 |
| Gender (Male) | 73 (89.02) | 67 (84.81) | 184 (83.26) | 60 (86.96) |
| Age (Years) | 63 (56-71) | 63 (56-72) | 69 (60-74) | 70 (60-74) |
| AGT1 (rs699) | ||||
| AA | 26 (31.71) | 25 (31.65) | 72 (32.58) | 22 (31.88) |
| AG | 34 (41.46) | 35 (44.3) | 101 (45.7) | 38 (55.07) |
| GG | 22 (26.83) | 19 (24.05) | 47 (21.27) | 9 (13.04) |
| NA | 0 (0) | 0 (0) | 1 (0.45) | 0 (0) |
| AGT2 (rs4762) | ||||
| AA | 5 (6.1) | 3 (3.8) | 5 (2.26) | 0 (0) |
| AG | 16 (19.51) | 10 (12.66) | 44 (19.91) | 15 (21.74) |
| GG | 61 (74.39) | 66 (83.54) | 172 (77.83) | 54 (78.26) |
| AHT (Yes) | 37 (45.12) | 36 (45.57) | 65 (29.41) | 45 (65.22) |
| Diabetes (Yes) | 22 (26.83) | 28 (35.44) | 61 (27.6) | 23 (33.33) |
| HBV (Yes) | 10 (12.2) | 6 (7.59) | 46 (20.81) | 12 (17.39) |
| HCV (Yes) | 54 (65.85) | 38 (48.1) | 111 (50.23) | 44 (63.77) |
| HIV (Yes) | 2 (2.44) | 1 (1.27) | 3 (1.36) | 0 (0) |
| Child-Pugh | ||||
| A: 5-6 | 70 (85.37) | 63 (79.75) | 207 (93.67) | 58 (84.06) |
| B: 7-9 | 10 (12.2) | 11 (13.93) | 14 (6.33) | 10 (14.49) |
| Not applicable | 2 (2.44) | 5 (6.33) | 0 (0) | 1 (1.45) |
| ECOG-PS (0) | 77 (93.9) | 74 (93.67) | 155 (70.14) | 69 (100) |
| Ascites (Yes) | 11 (13.41) | 9 (11.39) | 25 (11.31) | 14 (20.29) |
| Encephalopathy (Yes) | 0 (0) | 0 (0) | 11 (4.98) | 0 (0) |
| Extrahepatic spread (Yes) | 24 (29.27) | 27 (34.18) | 79 (35.75) | 23 (33.33) |
| Vascular Invasion (Yes) | 22 (26.83) | 26 (32.91) | 61 (27.6) | 31 (44.93) |
| BCLC (A1 or B / C) | 42 (51.22) / 40 (48.78) | 36 (45.57) / 43 (54.43) | 76 (34.39) / 145 (65.61) | 20 (28.99) / 49 (71.01) |
| Alpha-fetoprotein (ng/mL) | 20.5 (7-212.5) | 25 (8-228) | 100.5 (10-869) | 98 (5-1903) |
| Hemoglobin basal (g/dL) | 13.8 (12.95-14.95) | 13.1 (11.9-14.5) | 12.5 (11.2-14) | 13 (11.9-13.9) |
| Prothrombin time (%) | 88.3 (76.5-95.6) | 76 (65-88) | NA | 84.5 (76-100) |
| International normalized ratio | NA | NA | 1.1 (1-1.22) | 1.13 (1.03-1.24) |
| Total bilirubin (mg/dL) | 1 (0.8-1.6) | 1.1 (0.6-1.7) | 0.9 (0.72-1.3) | 0.95 (0.7-1.4) |
| AST (UI/L) | 78 (46-119) | 54 (34-84) | NA | 52 (35-80) |
| ALT (UI/L) | 72 (35-106.5) | 44 (25-65) | 43 (23-56) | 42 (32-55) |
| GGT (UI/L) | 134.5 (93.5-285.5) | 143 (83-264) | NA | 96 (48-204) |
| Albumin (mg/L) | 38.5 (35-43) | 40 (35-43) | 38 (35-40) | 3.6 (3.3-4) |
| Initial dosage of sorafenib (mg) | ||||
| 400 | 5 (6.1) | 2 (2.6) | 19 (8.6) | 0 (0) |
| 600 | 0 (0) | 0 (0) | 5 (2.26) | 0 (0) |
| 800 | 77 (93.9) | 77 (97.4) | 197 (89.14) | 69 (100) |
| SNP alleles (A/G) | Patients at risk | Events | Median OS (95%CI), months | P value (log-rank) | ||
| BCLC1cohort | 82 | 75 | 18.81 (14.76-23.58) | |||
| BCLC2 cohort | 79 | 47 | 18.32 (13.05-26.44) | |||
| Northern Italy cohort | 221 | 180 | 14.3 (11.84-17.99) | |||
| Naples cohort | 69 | 57 | 9.9 (7.69-12.82) | |||
| BCLC1 cohort | AGT1 (rs699) | 82 | 75 | 0.16 | ||
| AA | 26 | 23 | 18.73 (11.84-41.4) | |||
| AG | 34 | 33 | 18.43 (10.75-22.76) | |||
| GG | 22 | 19 | 18.81 (9.67-30.42) | |||
| AGT2 (rs4762) | 82 | 75 | 0.4 | |||
| AA | 5 | 5 | 41.34 (0.39-74.12) | |||
| AG | 16 | 15 | 13.95 (7.3-23.87) | |||
| GG | 61 | 55 | 19.11 (14.86-24.47) | |||
| BCLC2 cohort | AGT1 (rs699) | 79 | 47 | 0.15 | ||
| AA | 25 | 15 | 23.74 (7.46-26.5) | |||
| AG | 35 | 19 | 21.74 (11.15-33.77) | |||
| GG | 19 | 13 | 6.64 (3.42-30.29) | |||
| AGT2 (rs4762) | 79 | 47 | 0.3 | |||
| AA | 3 | 1 | NE (13.61-NE) | |||
| AG | 10 | 5 | 30.29 (3.88-32.69) | |||
| GG | 66 | 41 | 16.41 (8.78-23.74) | |||
| Northern Italy cohort | AGT1 (rs699) | 220 | 179 | 0.5 | ||
| AA | 72 | 58 | 13.58 (10.92-19.2) | |||
| AG | 101 | 83 | 17.59 (10.85-20.68) | |||
| GG | 47 | 38 | 12.43 (8.81-20.68) | |||
| AGT2 (rs4762) | 221 | 180 | 0.7 | |||
| AA | 5 | 2 | NE (1.94-NE) | |||
| AG | 44 | 36 | 14.3 (7.46-20.68) | |||
| GG | 172 | 142 | 14.9 (11.25-18.09) | |||
| Naples cohort | AGT1 (rs699) | 69 | 57 | 0.7 | ||
| AA | 22 | 19 | 12.66 (6.15-18.25) | |||
| AG | 38 | 31 | 8.32 (4.9-11.71) | |||
| GG | 9 | 7 | 10.95 (2.6-21.83) | |||
| AGT2 (rs4762) | 69 | 57 | 0.6 | |||
| AG | 15 | 11 | 9.8 (2.89-24.93) | |||
| GG | 54 | 46 | 10.1 (7.14-12.82) | |||
| BCLC1cohort | BCLC2cohort | Northern Italy cohort | Naples cohort | |
| Patients, n | 82 | 79 | 221 | 69 |
| Follow-up (mo) | 18.58 (10.33-34.17) | 13.05 (6.64-22.36) | 12.73 (6.05-25.88) | 9.87 (4.51-18.25) |
| Treatment duration (mo) | 9.06 (4.11-17.46) | 5.95 (2.14-13.52) | 8.52 (2.56-20.78) | 8.06 (3.72-16.97) |
| Adverse Events | ||||
| Gastrointestinal (Yes) | 35 (42.68) | 27 (34.18) | 23 (10.41) | 38 (55.07) |
| Dermatologic (Yes) | 42 (51.22) | 28 (35.44) | 32 (14.48) | 27 (39.13) |
| Early Dermatologic (Yes) | 33 (40.24) | 22 (27.85) | 28 (12.67) | 25 (36.23) |
| Performance status deterioration (Yes) | 44 (53.66) | 46 (58.23) | 53 (23.98) | 0 (0) |
| Cardiovascular (Yes) | 18 (21.95) | 14 (17.72) | 16 (7.24) | 16 (23.19) |
| Dermatologic and Cardiovascular simultaneously (Yes) | 7 (8.54) | 5 (6.33) | 0 (0) | 10 (14.49) |
| Other (Yes) | 48 (58.54) | 34 (43.04) | 45 (20.36) | 65 (94.2) |
| Death (Yes) | 75 (91.46) | 47 (59.49) | 180 (81.44) | 57 (82.61) |
| Cause of death | ||||
| HCC | 74 (98.67) | 46 (97.87) | 118 (65.56) | 48 (84.21) |
| Not HCC related | 0 (0) | 1 (2.13) | 58 (32.22) | 9 (15.79) |
| Others1 | 1 (1.33) | 0 (0) | 4 (2.22) | 0 (0) |
BCLC1 cohort: All but 2 (2.4%) patients were cirrhotic. A total of 54 (65.9%) patients had Hepatitis C Virus (HCV) and 10 (12.2%) had Hepatitis B Virus (HBV). Ninety-three percent of patients were asymptomatic (ECOG-PS 0) and 40 (48.8%) were BCLC B that failed or had a contraindication to loco-regional treatment, 70 (85.4%) were Child-Pugh class A. Twenty-two (26.8%) had vascular invasion, and 24 (29.3%) had extra-hepatic spread. AHT was present in 45.1% of patients and diabetes in 26.8%. Seventy-seven patients (93.9%) started sorafenib treatment at 800 mg.
BCLC2 cohort: All but 5 (6.3%) patients were cirrhotic. A total of 38 (48.1%) patients had HCV and 6 (7.6%) had HBV. Ninety-three percent of patients were asymptomatic (ECOG-PS 0) and 36 (45.6%) were BCLC B that failed or had a contraindication to loco-regional treatment, 63 (79.8%) were Child-Pugh class A. Twenty-six (32.9%) had vascular invasion, and 27 (34.2%) had extra-hepatic spread. AHT was present in 45.6% of patients and diabetes in 35.4%. Seventy-seven patients (97.4%) started sorafenib treatment at 800 mg.
Northern Italy cohort: All patients were cirrhotic. A total of 111 (50.2%) patients had HCV and 46 (20.8%) had HBV. Seventy percent of patients were asymptomatic (ECOG-PS 0) and 76 (34.4%) were BCLC B that failed or had a contraindication to loco-regional treatment, 207 (93.7%) were Child-Pugh class A. Sixty-one (27.6%) had vascular invasion, and 79 (35.8%) had extra-hepatic spread. AHT was present in 29.4% of patients and diabetes in 27.6%. One hundred ninety-seven patients (89.1%) started sorafenib treatment at 800 mg.
Naples cohort: All but 1 (1.5%) patient were cirrhotic. A total of 44 (63.7%) patients had HCV and 12 (17.4%) had HBV. All patients were asymptomatic (ECOG-PS 0) and 20 (29%) were BCLC B that failed or had a contraindication to loco-regional treatment, 58 (84.1%) were Child-Pugh class A. Thirty-one (44.9%) had vascular invasion, and 23 (33.3%) had extra-hepatic spread. AHT was present in 65.2% of patients and diabetes in 33.3%. All patients started sorafenib treatment at 800 mg.
The rate of DAEs at any time point in the BCLC1, BCLC2, Northern Italy and Naples cohorts were 51.2 %, 35.4%, 14.5% and 39.1%, respectively (Table 3). The incidence of eDAEs in the BCLC1 cohort was 40.2% and was 27.8%, 12.7% and 36.2% in the BCLC2, Northern Italy and Naples cohorts, respectively.
The distribution of patients with a history of diabetes and AHT who did or did not develop eDAEs or DAEs in each cohort and the association between DAEs and AHT are summarized in Supple
The association between DAEs and a history of AHT was statistically significant in the BCLC1 cohort, with a HR = 1.96 (95%CI: 1.05-3.65; P value = 0.04) and confirmed when all patients were analyzed as a unique cohort with a HR = 1.61 (95%CI: 1.14-2.28; P value = 0.007).
BCLC1 cohort: The median follow-up was 18.6 mo (IQR: 10.3-34.2) and 75 (91.5%) patients died. Ninety-eight percent of deaths were due to HCC-related causes. The median treatment duration and OS were 9.1 (IQR: 4.1-17.5) and 18.8 mo (95%CI: 14.7-23.6), respectively.
BCLC2 cohort: The median follow-up was 13.1 mo (IQR: 6.6-22.4) and 47 (59.5%) patients died. Ninety-seven percent of deaths were due to HCC-related causes. The median treatment duration and OS were 5.9 (IQR: 2.1-13.5) and 18.3 mo (95%CI: 13.1-26.4), respectively.
Northern Italy cohort: The median follow-up was 12.7 mo (IQR: 6.1-25.9) and 180 (81.4%) patients died. Sixty-five percent of deaths were due to HCC-related causes. The median treatment duration and OS were 8.5 (IQR: 2.6-20.8) and 14.3 mo (95%CI: 11.8-18), respectively.
Naples cohort: The median follow-up was 9.9 mo (IQR: 4.5-18.3) and 57 (82.6%) patients died. Eighty-four percent of deaths were due to HCC-related causes. The median treatment duration and OS were 8.1 (IQR: 3.7-17) and 9.9 mo (95%CI: 7.7-12.8), respectively.
Using a landmark timepoint of 60 (+7) days and excluding 17 patients with less than 60 (+7) days of follow-up, the median OS in eDAE and in non-eDAE patients was 21.6 mo (95%CI: 12.7-28.2) and 14.8 mo (95%CI: 9.9-17.6) in BCLC1, 19.5 mo (95%CI: 8-24.2) and 14.2 mo (95%CI: 8.9-30.5) in BCLC2, 15.9 mo (95%CI: 8.3-40.6) and 12.1 mo (95%CI: 9.6-16.6) in the Northern Italy cohort, 12.4 mo (95%CI: 7.86-21.14) and 6.8 mo (95%CI: 2.7-8.7) in the Naples cohort, respectively.
BCLC1 cohort: Supplementary Table 1 describes the assessed SNPs in this cohort. Of all SNPs analyzed, only the AGT1 (rs699) AA genotype had a significant estimated increase in the probability of eDAE with a HR = 2.31 (95%CI: 1.03-5.14; P value = 0.04; AA vs AG) in the univariate model and a HR = 2.3 (95%CI: 1.02-5.16; P value = 0.04; AA vs AG) in the multivariate model (Table 4). For DAEs at any time point, AGT1 (rs699) AA genotype showed a significant estimated increase in the probability of DAEs with a HR = 2.7 (95%CI: 1.27-5.75; P value = 0.01; AA vs AG) in the univariate model and a HR = 2.68 (95%CI: 1.25-5.77; P value = 0.01; AA vs AG) in the multivariate model. No other polymorphism showed a significant association with general AEs or specifically DAE or eDAE development in the BCLC1 cohort.
| Event | Centre | AGT1(rs699) | HR (95%CI) | P value | HR (95%CI) adjusted by BCLC + ECOG-PS | P value | HR (95%CI) adjusted by BCLC + ECOG-PS + AHT + DM | P value | HR (95%CI) adjusted for AHT + DM | P value | HR (95%CI) adjusted for DM | P value | HR (95%CI) adjusted for AHT | P value |
| eDAE | BCLC1 cohort | AA vs AG | 2.31 (1.03-5.14) | 0.04 | 2.3 (1.02-5.16) | 0.04 | 2.34 (1.02-5.37) | 0.04 | 2.33 (1.03-5.24) | 0.04 | 2.45 (1.1-5.5) | 0.03 | 2.24 (1-5.03) | 0.049 |
| AA vs GG | 1.68 (0.71-3.97) | 0.2 | 1.69 (0.71-4) | 0.2 | 1.64 (0.69-3.93) | 0.3 | 1.65 (0.69-3.92) | 0.3 | 1.75 (0.74-4.13) | 0.2 | 1.62 (0.68-3.87) | 0.3 | ||
| AG vs GG | 0.73 (0.29-1.85) | 0.5 | 0.73 (0.29-1.89) | 0.5 | 0.7 (0.27-1.82) | 0.5 | 0.71 (0.28-1.79) | 0.5 | 0.71 (0.28-1.8) | 0.5 | 0.72 (0.29-1.84) | 0.5 | ||
| BCLC2 cohort | AA vs AG | 0.66 (0.25-1.76) | 0.4 | 0.63 (0.24-1.7) | 0.4 | 0.71 (0.26-1.93) | 0.5 | 0.72 (0.27-1.93) | 0.5 | 0.72 (0.27-1.91) | 0.5 | 0.68 (0.25-1.83) | 0.5 | |
| AA vs GG | 1.13 (0.32-4.01) | 0.9 | 1.08 (0.3-3.84) | 0.9 | 1.35 (0.37-4.95) | 0.7 | 1.36 (0.38-4.9) | 0.7 | 1.32 (0.37-4.72) | 0.7 | 1.13 (0.32-4) | 0.9 | ||
| AG vs GG | 1.71 (0.55-5.3) | 0.4 | 1.7 (0.55-5.28) | 0.4 | 1.89 (0.6-5.91) | 0.3 | 1.89 (0.6-5.9) | 0.3 | 1.85 (0.6-5.74) | 0.3 | 1.66 (0.53-5.17) | 0.4 | ||
| Northern Italy cohort | AA vs AG | 0.8 (0.33-1.95) | 0.6 | 0.75 (0.3-1.86) | 0.5 | 1.02 (0.4-2.61) | 0.9 | 0.96 (0.39-2.36) | 0.9 | 0.83 (0.34-2.02) | 0.7 | 0.91 (0.37-2.23) | 0.8 | |
| AA vs GG | 0.9 (0.31-2.6) | 0.8 | 0.71 (0.24-2.1) | 0.5 | 0.96 (0.31-2.98) | 0.9 | 1.22 (0.4-3.73) | 0.7 | 0.96 (0.33-2.8) | 0.9 | 1.12 (0.37-3.36) | 0.8 | ||
| AG vs GG | 1.12 (0.42-3.01) | 0.8 | 0.95 (0.35-2.58) | 0.9 | 0.94 (0.33-2.69) | 0.9 | 1.27 (0.46-3.49) | 0.7 | 1.15 (0.43-3.12) | 0.8 | 1.23 (0.45-3.34) | 0.7 | ||
| Naples cohort | AA vs AG | 1.26 (0.54-2.95) | 0.6 | 1.21 (0.51-2.86) | 0.7 | 1.35 (0.56-3.27) | 0.5 | 1.36 (0.57-3.25) | 0.5 | 1.23 (0.52-2.93) | 0.6 | 1.44 (0.61-3.39) | 0.4 | |
| AA vs GG | 1.26 (0.34-4.66) | 0.7 | 1.18 (0.31-4.43) | 0.8 | 1.33 (0.35-5) | 0.7 | 1.34 (0.36-4.96) | 0.7 | 1.27 (0.34-4.68) | 0.7 | 1.35 (0.37-5) | 0.7 | ||
| AG vs GG | 1 (0.28-3.51) | 0.9 | 0.97 (0.28-3.43) | 0.9 | 0.98 (0.28-3.49) | 0.9 | 0.99 (0.28-3.49) | 0.9 | 1.03 (0.29-3.65) | 0.9 | 0.94 (0.27-3.3) | 0.9 | ||
| BCLC2 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 0.87 (0.52-1.47) | 0.6 | 0.85 (0.51-1.43) | 0.5 | 0.84 (0.5-1.41) | 0.5 | 0.85 (0.51-1.43) | 0.6 | 0.87 (0.52-1.47) | 0.6 | 0.85 (0.51-1.43) | 0.6 | |
| AA vs GG | 1.05 (0.54-2.04) | 0.9 | 0.95 (0.49-1.86) | 0.9 | 0.92 (0.47-1.81) | 0.8 | 1.01 (0.52-1.97) | 0.9 | 1.05 (0.54-2.04) | 0.9 | 1.01 (0.52-1.97) | 0.9 | ||
| AG vs GG | 1.2 (0.65-2.22) | 0.6 | 1.12 (0.61-2.08) | 0.7 | 1.1 (0.59-2.05) | 0.8 | 1.18 (0.64-2.18) | 0.6 | 1.2 (0.65-2.22) | 0.6 | 1.18 (0.64-2.18) | 0.6 | ||
| BCLC1 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 1.35 (0.84-2.17) | 0.2 | 1.35 (0.84-2.18) | 0.2 | 1.33 (0.82-2.15) | 0.2 | 1.31 (0.81-2.11) | 0.3 | 1.35 (0.84-2.18) | 0.2 | 1.3 (0.81-2.1) | 0.3 | |
| AA vs GG | 1.19 (0.67-2.12) | 0.6 | 1.13 (0.6-2.01) | 0.7 | 1.08 (0.6-1.93) | 0.8 | 1.1 (0.61-1.97) | 0.8 | 1.19 (0.67-2.12) | 0.6 | 1.09 (0.61-1.96) | 0.8 | ||
| AG vs GG | 0.88 (0.5-1.55) | 0.7 | 0.83 (0.47-1.48) | 0.5 | 0.81 (0.46-1.43) | 0.5 | 0.84 (0.48-1.48) | 0.6 | 0.88 (0.5-1.55) | 0.7 | 0.84 (0.48-1.48) | 0.6 | ||
| BCLC1cohort + BCLC2 cohort + Naples cohort | AA vs AG | 1.32 (0.81-2.15) | 0.3 | 1.29 (0.79-2.11) | 0.3 | 1.3 (0.79-2.12) | 0.3 | 1.31 (0.81-2.14) | 0.3 | 1.33 (0.82-2.17) | 0.3 | 1.31 (0.8-2.13) | 0.3 | |
| AA vs GG | 1.4 (0.75-2.6) | 0.3 | 1.38 (0.7-2.57) | 0.3 | 1.44 (0.77-2.69) | 0.3 | 1.45 (0.78-2.7) | 0.2 | 1.46 (0.79-2.72) | 0.2 | 1.38 (0.74-2.57) | 0.3 | ||
| AG vs GG | 1.06 (0.58-1.94) | 0.9 | 1.06 (0.58-1.95) | 0.9 | 1.11 (0.6-2.03) | 0.8 | 1.1 (0.6-2.02) | 0.8 | 1.1 (0.6-2.01) | 0.8 | 1.06 (0.58-1.94) | 0.9 | ||
| DAE | BCLC1 cohort | AA vs AG | 2.7 (1.27-5.75) | 0.01 | 2.68 (1.25-5.77) | 0.01 | 2.52 (1.16-5.47) | 0.02 | 2.6 (1.21-5.57) | 0.01 | 2.82 (1.32-6.06) | 0.008 | 2.5 (1.17-5.35) | 0.02 |
| AA vs GG | 1.26 (0.62-2.58) | 0.5 | 1.24 (0.61-2.55) | 0.6 | 1.11 (0.53-2.31) | 0.8 | 1.13 (0.55-2.35) | 0.8 | 1.3 (0.63-2.66) | 0.5 | 1.12 (0.54-2.32) | 0.8 | ||
| AG vs GG | 0.47 (0.21-1.05) | 0.06 | 0.46 (0.2-1.06) | 0.07 | 0.44 (0.19-1.01) | 0.053 | 0.44 (0.19-0.98) | 0.045 | 0.46 (0.2-1.03) | 0.06 | 0.45 (0.2-1.01) | 0.052 | ||
| BCLC2 cohort | AA vs AG | 0.98 (0.43-2.2) | 0.9 | 0.94 (0.42-2.13) | 0.9 | 0.99 (0.43-2.26) | 0.9 | 1.01 (0.45-2.3) | 0.9 | 1.03 (0.45-2.32) | 0.9 | 0.95 (0.42-2.16) | 0.9 | |
| AA vs GG | 1.89 (0.59-6.04) | 0.3 | 1.78 (0.55-5.76) | 0.3 | 2.08 (0.63-6.85) | 0.2 | 2.18 (0.67-7.03) | 0.19 | 2.08 (0.65-6.66) | 0.2 | 1.88 (0.59-6.01) | 0.3 | ||
| AG vs GG | 1.94 (0.64-5.9) | 0.2 | 1.89 (0.62-5.77) | 0.3 | 2.12 (0.69-6.49) | 0.19 | 2.15 (0.7-6.57) | 0.18 | 2.02 (0.66-6.15) | 0.2 | 1.98 (0.65-6.05) | 0.2 | ||
| Northern Italy cohort | AA vs AG | 0.89 (0.39-2.06) | 0.8 | 0.85 (0.37-1.98) | 0.7 | 1.01 (0.42-2.41) | 0.9 | 1 (0.42-2.33) | 0.9 | 0.91 (0.39-2.11) | 0.8 | 0.95 (0.41-2.22) | 0.9 | |
| AA vs GG | 0.62 (0.25-1.57) | 0.3 | 0.54 (0.21-1.37) | 0.2 | 0.6 (0.23-1.6) | 0.3 | 0.74 (0.28-1.92) | 0.5 | 0.64 (0.25-1.62) | 0.4 | 0.7 (0.27-1.79) | 0.5 | ||
| AG vs GG | 0.7 (0.3-1.62) | 0.3 | 0.63 (0.27-1.48) | 0.2 | 0.6 (0.25-1.43) | 0.2 | 0.74 (0.32-1.72) | 0.5 | 0.71 (0.3-1.63) | 0.4 | 0.73 (0.31-1.69) | 0.5 | ||
| Naples cohort | AA vs AG | 1.29 (0.57-2.92) | 0.5 | 1.23 (0.54-2.81) | 0.6 | 1.35 (0.58-3.15) | 0.5 | 1.38 (0.6-3.17) | 0.5 | 1.23 (0.54-2.81) | 0.6 | 1.49 (0.66-3.4) | 0.3 | |
| AA vs GG | 1.38 (0.38-5.03) | 0.6 | 1.28 (0.35-4.73) | 0.7 | 1.45 (0.39-5.36) | 0.6 | 1.49 (0.41-5.41) | 0.6 | 1.39 (0.38-5.05) | 0.6 | 1.51 (0.41-5.51) | 0.5 | ||
| AG vs GG | 1.07 (0.31-3.72) | 0.9 | 1.04 (0.3-3.62) | 0.9 | 1.08 (0.31-3.77) | 0.9 | 1.08 (0.31-3.79) | 0.9 | 1.13 (0.32-3.96) | 0.9 | 1.01 (0.29-3.52) | 0.9 | ||
| BCLC2 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 1 (0.62-1.61) | 0.9 | 0.98 (0.61-1.57) | 0.9 | 0.95 (0.59-1.54) | 0.9 | 0.97 (0.6-1.56) | 0.9 | 1.01 (0.63-1.62) | 0.9 | 0.96 (0.6-1.55) | 0.9 | |
| AA vs GG | 1.13 (0.61-2.08) | 0.7 | 1.04 (0.56-1.92) | 0.9 | 0.98 (0.53-1.81) | 0.9 | 1.05 (0.57-1.95) | 0.9 | 1.12 (0.61-2.07) | 0.7 | 1.05 (0.57-1.95) | 0.9 | ||
| AG vs GG | 1.13 (0.63-2) | 0.7 | 1.06 (0.59-1.89) | 0.8 | 1.02 (0.57-1.83) | 0.9 | 1.09 (0.61-1.94) | 0.8 | 1.12 (0.63-1.99) | 0.7 | 1.09 (0.61-1.95) | 0.8 | ||
| BCLC1 cohort + Naples cohort +Northern Italy cohort | AA vs AG | 1.43 (0.91-2.24) | 0.12 | 1.43 (0.91-2.24) | 0.12 | 1.39 (0.88-2.19) | 0.15 | 1.36 (0.87-2.14) | 0.18 | 1.44 (0.92-2.26) | 0.11 | 1.35 (0.86-2.12) | 0.19 | |
| AA vs GG | 0.94 (0.57-1.56) | 0.8 | 0.9 (0.54-1.51) | 0.7 | 0.82 (0.49-1.38) | 0.5 | 0.83 (0.49-1.39) | 0.5 | 0.94 (0.57-1.57) | 0.8 | 0.82 (0.49-1.38) | 0.5 | ||
| AG vs GG | 0.66 (0.4-1.09) | 0.1 | 0.63 (0.38-1.05 | 0.08 | 0.59 (0.36-0.99) | 0.04 | 0.61 (0.37-1.01) | 0.052 | 0.66 (0.4-1.08) | 0.1 | 0.61 (0.37-1.01) | 0.053 | ||
| BCLC1 cohort + BCLC2 cohort + Naples cohort | AA vs AG | 1.54 (0.98-2.41) | 0.06 | 1.49 (0.95-2.34) | 0.08 | 1.48 (0.94-2.32) | 0.09 | 1.52 (0.97-2.37) | 0.07 | 1.55 (0.99-2.43) | 0.055 | 1.5 (0.96-2.35) | 0.07 | |
| AA vs GG | 1.35 (0.78-2.32) | 0.3 | 1.3 (0.75-2.25) | 0.3 | 1.32 (0.76-2.28) | 0.3 | 1.35 (0.78-2.33) | 0.3 | 1.39 (0.81-2.4) | 0.2 | 1.3 (0.76-2.24) | 0.3 | ||
| AG vs GG | 0.88 (0.51-1.51) | 0.6 | 0.87 (0.51-1.5) | 0.6 | 0.89 (0.52-1.54) | 0.7 | 0.89 (0.52-1.54) | 0.7 | 0.9 (0.52-1.54) | 0.7 | 0.87 (0.5-1.49) | 0.6 |
Allele distributions of AGT1 (rs699) and AGT2 (rs4762) are summarized in Table 1. There were no significant differences between the included cohorts (P value 0.5 and 0.2 for AGT1 rs699 and AGT2 rs4762, respectively). Thus, the present cohorts are comparable in terms of genetic variants.
Tables 4 and 5 describe the Cox regression models for eDAE and DAE development by AGT1 (rs699) and AGT2 (rs4762), respectively. The results of the BCLC1 cohort are mentioned above.
| Event | Center | AGT2(rs4762) | HR (95%CI) | P value | HR (95%CI) adjusted for BCLC + ECOG-PS | P value | HR (95%CI) adjusted for BCLC + ECOG-PS + AHT + DM | P value | HR (95%CI) adjusted for AHT + DM | P value | HR (95%CI) adjusted for DM | P value | HR (95%CI) adjusted for AHT | P value |
| eDAE | BCLC1 cohort | AA vs AG | 1.14 (0.22-5.89) | 0.9 | 0.98 (0.19-5.12) | 0.9 | 0.97 (0.18-5.04) | 0.9 | 1.09 (0.21-5.64) | 0.9 | 1.15 (0.22-5.95) | 0.9 | 1.11 (0.21-5.72) | 0.9 |
| AA vs GG | 0.84 (0.2-3.53) | 0.8 | 0.73 (0.17-3.15) | 0.7 | 0.71 (0.16-3.1) | 0.7 | 0.81 (0.19-3.4) | 0.8 | 0.8 (0.19-3.39) | 0.8 | 0.84 (0.2-3.54) | 0.8 | ||
| AG vs GG | 0.73 (0.28-1.91) | 0.5 | 0.74 (0.28-1.94) | 0.5 | 0.74 (0.28-1.97) | 0.6 | 0.74 (0.28-1.94) | 0.6 | 0.7 (0.27-1.82) | 0.5 | 0.76 (0.29-1.98) | 0.6 | ||
| BCLC2 cohort | AA vs AG | 3.71 (0.62-22.39) | 0.2 | 3.52 (0.58-21.5) | 0.2 | 4.8 (0.74-31.28) | 0.1 | 4.81 (0.74-31.24) | 0.1 | 4.78 (0.76-29.88) | 0.09 | 4.46 (0.7-28.35) | 0.11 | |
| AA vs GG | 4.43 (1.01-19.39) | 0.048 | 4.24 (0.95-19.06) | 0.06 | 6.14 (1.28-29.55) | 0.02 | 6.28 (1.32-29.95) | 0.02 | 6.25 (1.35-28.89) | 0.02 | 5.34 (1.15-24.86) | 0.03 | ||
| AG vs GG | 1.19 (0.35-4.08) | 0.8 | 1.21 (0.35-4.15) | 0.8 | 1.28 (0.37-4.45) | 0.7 | 1.31 (0.38-4.47) | 0.7 | 1.31 (0.38-4.47) | 0.7 | 1.2 (0.35-4.08) | 0.8 | ||
| Northern Italy cohort | AA vs AG | 2.72 (0.57-13.1) | 0.2 | 3.21 (0.64-15.99) | 0.15 | 5.61 (1.01-31.12) | 0.048 | 3.43 (0.69-16.96) | 0.13 | 2.69 (0.56-12.97) | 0.2 | 3.2 (0.66-15.6) | 0.15 | |
| AA vs GG | 4.54 (1.05-19.64) | 0.04 | 5.15 (1.17-22.63) | 0.03 | 8.51 (1.78-40.54) | 0.007 | 5.51 (1.25-24.33) | 0.02 | 4.72 (1.09-20.48) | 0.04 | 4.93 (1.13-21.41) | 0.03 | ||
| AG vs GG | 1.67 (0.69-4.02) | 0.3 | 1.6 (0.66-3.9) | 0.3 | 1.52 (0.6-3.82) | 0.4 | 1.61 (0.66-3.9) | 0.3 | 1.75 (0.73-4.24) | 0.2 | 1.54 (0.63-3.73) | 0.3 | ||
| Naples cohort | AG vs GG | 1.2 (0.48-3.01) | 0.7 | 1.2 (0.48-3.02) | 0.7 | 1.25 (0.5-3.15) | 0.6 | 1.26 (0.5-3.16) | 0.6 | 1.2 (0.48-3) | 0.7 | 1.29 (0.51-3.23) | 0.6 | |
| BCLC2 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 2.76 (0.92-8.27) | 0.07 | 2.95 (0.97-9.84) | 0.06 | 2.78 (0.9-8.56) | 0.07 | 2.61 (0.87-7.86) | 0.09 | 2.75 (0.92-8.25) | 0.07 | 2.61 (0.87-7.86) | 0.09 | |
| AA vs GG | 3.5 (1.27-9.67) | 0.02 | 3.8 (1.36-10.58) | 0.01 | 3.67 (1.31-10.3) | 0.01 | 3.39 (1.22-9.37) | 0.02 | 3.5 (1.27-9.66) | 0.02 | 3.38 (1.22-9.37) | 0.02 | ||
| AG vs GG | 1.27 (0.73-9.67) | 0.4 | 1.29 (0.74-2.25) | 0.4 | 1.32 (0.75-2.32) | 0.3 | 1.3 (0.74-2.27) | 0.4 | 1.27 (0.73-2.22) | 0.4 | 1.3 (0.74-2.27) | 0.4 | ||
| BCLC1 cohort + Naples cohort +Northern Italy cohort | AA vs AG | 1.66 (0.65-4.9) | 0.4 | 1.63 (0.55-4.85) | 0.4 | 1.53 (0.51-4.57) | 0.5 | 1.54 (0.52-4.57) | 0.4 | 1.66 (0.56-4.9) | 0.4 | 1.54 (0.52-4.57) | 0.4 | |
| AA vs GG | 1.83 (0.67-5.03) | 0.2 | 1.73 (0.63-4.77) | 0.3 | 1.7 (0.62-4.69) | 0.3 | 1.8 (0.65-4.94) | 0.3 | 1.85 (0.67-5.08) | 0.2 | 1.79 (0.65-4.93) | 0.3 | ||
| AG vs GG | 1.1 (0.65-1.86) | 0.7 | 1.06 (0.63-1.81) | 0.8 | 1.11 (0.65-1.9) | 0.7 | 1.17 (0.69-1.97) | 0.6 | 1.11 (0.66-1.88) | 0.7 | 1.16 (0.69-1.97) | 0.6 | ||
| BCLC1 cohort + BCLC2 cohort + Naples cohort | AA vs AG | 1.67 (0.55-5.09) | 0.4 | 1.6 (0.53-4.87) | 0.4 | 1.61 (0.53-4.92) | 0.4 | 1.66 (0.55-5.06) | 0.4 | 1.71 (0.56-5.19) | 0.4 | 1.63 (0.54-4.95) | 0.4 | |
| AA vs GG | 1.7 (0.62-4.67) | 0.3 | 1.67 (0.61-4.59) | 0.3 | 1.68 (0.61-4.63) | 0.3 | 1.7 (0.62-4.67) | 0.3 | 1.7 (0.62-4.67) | 0.3 | 1.68 (0.61-4.62) | 0.3 | ||
| AG vs GG | 1.01 (0.57-1.81) | 0.9 | 1.04 (0.58-1.86) | 0.9 | 1.04 (0.58-1.86) | 0.9 | 1.02 (0.57-1.82) | 0.9 | 0.99 (0.56-1.78) | 0.9 | 1.03 (0.58-1.84) | 0.9 | ||
| DAE | BCLC1 cohort | AA vs AG | 2.8 (0.78-10.01) | 0.1 | 2.45 (0.68-8.81) | 0.2 | 2.73 (0.74-9.99) | 0.13 | 3.09 (0.85-11.2) | 0.09 | 2.85 (0.79-10.22) | 0.11 | 2.86 (0.8-10.28) | 0.11 |
| AA vs GG | 1.82 (0.64-5.16) | 0.3 | 1.61 (0.56-4.64) | 0.4 | 1.89 (0.64-5.57) | 0.2 | 2.12 (0.74-6.1) | 0.16 | 1.79 (0.63-5.08) | 0.3 | 2.03 (0.71-5.78) | 0.19 | ||
| AG vs GG | 0.65 (0.27-1.56) | 0.3 | 0.66 (0.27-1.59) | 0.4 | 0.69 (0.28-1.72) | 0.4 | 0.69 (0.28-1.68) | 0.4 | 0.63 (0.26-1.52) | 0.3 | 0.71 (0.29-1.71) | 0.4 | ||
| BCLC2 cohort | AA vs AG | 3.83 (0.64-23.05) | 0.1 | 3.71 (0.61-22.68) | 0.2 | 3.91 (0.62-24.73) | 0.15 | 4.05 (0.65-25.33) | 0.14 | 4.49 (0.73-27.55) | 0.1 | 3.79 (0.61-23.44) | 0.15 | |
| AA vs GG | 3.22 (0.75-13.76) | 0.1 | 3.27 (0.74-14.38) | 0.1 | 3.74 (0.82-17.15) | 0.09 | 3.7 (0.82-16.76) | 0.09 | 4.04 (0.91-18) | 0.07 | 3.18 (0.72-14.13) | 0.13 | ||
| AG vs GG | 0.84 (0.25-2.8) | 0.8 | 0.88 (0.26-2.96) | 0.8 | 0.96 (0.28-3.24) | 0.9 | 0.92 (0.27-3.06) | 0.9 | 0.9 (0.27-3.01) | 0.9 | 0.84 (0.25-2.8) | 0.8 | ||
| Northern Italy cohort | AA vs AG | 2.85 (0.59-13.73) | 0.2 | 3.28 (0.66-16.21) | 0.1 | 4.71 (0.89-24.91) | 0.07 | 3.4 (0.69-16.77) | 0.13 | 2.83 (0.59-13.64) | 0.2 | 3.13 (0.65-15.21) | 0.16 | |
| AA vs GG | 3.68 (0.86-15.63) | 0.08 | 4.15 (0.96-17.87) | 0.06 | 5.97 (1.32-27.01) | 0.02 | 4.41 (1.02-19.03) | 0.046 | 3.97 (0.93-16.94) | 0.06 | 3.8 (0.89-16.16) | 0.07 | ||
| AG vs GG | 1.29 (0.55-3.01) | 0.6 | 1.26 (0.54-2.96) | 0.6 | 1.27 (0.53-3.05) | 0.6 | 1.3 (0.55-3.05) | 0.6 | 1.4 (0.6-3.29) | 0.4 | 1.21 (0.52-2.84) | 0.7 | ||
| Naples cohort | AG vs GG | 1.12 (0.45-2.77) | 0.8 | 1.11 (0.45-2.76) | 0.8 | 1.12 (0.45-2.79) | 0.8 | 1.13 (0.46-2.82) | 0.8 | 1.12 (0.45-2.77) | 0.9 | 1.16 (0.47-2.88) | 0.8 | |
| BCCL2 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 2.79 (0.93-8.35) | 0.07 | 3.04 (1-9.21) | 0.049 | 2.72 (0.88-8.34) | 0.08 | 2.54 (0.84-7.63) | 0.1 | 2.74 (0.92-8.21) | 0.07 | 2.56 (0.85-7.7) | 0.09 | |
| AA vs GG | 2.96 (1.08-8.13) | 0.03 | 3.27 (1.18-9.05) | 0.02 | 3.07 (1.1-8.56) | 0.03 | 2.81 (1.02-7.73) | 0.045 | 2.94 (1.07-8.07) | 0.04 | 2.83 (1.03-7.78) | 0.04 | ||
| AG vs GG | 1.06 (0.62-1.83) | 0.8 | 1.07 (0.62-1.86) | 0.8 | 1.13 (0.65-1.96) | 0.7 | 1.11 (0.64-1.92) | 0.7 | 1.07 (0.62-1.85) | 0.8 | 1.11 (0.64-1.91) | 0.7 | ||
| BCLC1 cohort + Naples cohort + Northern Italy cohort | AA vs AG | 2.82 (1.13-7.07) | 0.03 | 2.9 (1.15-7.32) | 0.02 | 2.7 (1.06-6.84) | 0.04 | 2.66 (1.06-6.69) | 0.04 | 2.81 (1.12-7.05) | 0.03 | 2.68 (1.07-6.74) | 0.04 | |
| AA vs GG | 2.86 (1.24-6.58) | 0.01 | 2.84 (1.23-6.54) | 0.01 | 2.85 (1.24-6.57) | 0.01 | 2.94 (1.28-6.77) | 0.01 | 2.91 (1.27-6.7) | 0.01 | 2.94 (1.28-6.77) | 0.01 | ||
| AG vs GG | 1.01 (0.61-1.68) | 0.9 | 0.98 (0.59-1.63) | 0.9 | 1.06 (0.63-1.77) | 0.8 | 1.1 (0.67-1.83) | 0.7 | 1.04 (0.63-1.71) | 0.9 | 1.1 (0.66-1.82) | 0.7 | ||
| BCLC1 cohort + BCLC2 cohort + Naples cohort | AA vs AG | 2.94 (1.14-7.6) | 0.03 | 2.85 (1.1-7.39) | 0.03 | 3.12 (1.2-8.14) | 0.02 | 3.21 (1.23-8.34) | 0.02 | 3.05 (1.18-7.9) | 0.02 | 2.9 (1.12-7.5) | 0.03 | |
| AA vs GG | 2.49 (1.08-5.73) | 0.03 | 2.48 (1.08-5.72) | 0.03 | 2.73 (1.18-6.32) | 0.02 | 2.75 (1.19-6.34) | 0.02 | 2.54 (1.1-5.85) | 0.03 | 2.51 (1.09-5.77) | 0.03 | ||
| AG vs GG | 0.85 (0.49-1.48) | 0.6 | 0.87 (0.5-1.52) | 0.6 | 0.87 (0.5-1.53) | 0.7 | 0.86 (0.49-1.5) | 0.6 | 0.83 (0.48-1.45) | 0.5 | 0.87 (0.5-1.51) | 0.6 |
BCLC2 cohort: The AGT1 (rs699) did not show a significant association with DAEs. By contrast, the AGT2 (rs4762) AA genotype was associated with a significant increased risk of eDAE with a HR = 4.43 (95%CI: 1.01-19.39; P value = 0.048; AA vs GG) in the univariate analysis, and showed a trend in the multivariate model with a HR = 4.24 (95%CI: 0.95-19.06]; P value = 0.06; AA vs GG), Table 5.
Northern Italy cohort: In this cohort, the AGT2 (rs4762) AA genotype showed a statistically significant increased probability of eDAE both in the univariate analysis (HR = 4.54 [95%CI: 1.05-19.64]; P value = 0.04; AA vs GG) and in the multivariate analysis (HR = 5.15 [95%CI: 1.17-22.63]; P value = 0.03; AA vs GG).
Naples cohort: In the Naples cohort, none of the SNPs showed a significant effect on DAE or eDAE development.
The results in the individual cohorts suggested that the inconclusive results obtained in the BCLC and Naples cohorts could be due to a limited sample size. Thus, we combined these cohorts into a single cohort that would match the Northern Italy sample size.
This analysis showed that AGT2 (rs4762) was significantly associated with DAE development with a HR = 2.94 (95%CI: 1.14-7.6; P value = 0.03; AA vs AG) and HR = 2.49 (95%CI: 1.08-5.73; P value = 0.03; AA vs GG) in univariate models, and HR = 2.85 (95%CI: 1.1-7.39; P value = 0.03; AA vs AG) and HR = 2.48 (95%CI: 1.08-5.72; P value = 0.03; AA vs GG) in multivariate models (Table 5).
Table 5 shows the multivariate analyses adjusted for baseline BCLC stage, ECOG-PS, diabetes and AHT in the same model, considering diabetes and AHT together and each one separately. The multivariate analysis adjusted for baseline BCLC stage, ECOG-PS, diabetes and AHT showed a statistically significant increased risk in the probability of eDAE in patients harboring AGT2 (rs4762) AA genotype in the Northern Italy cohort (HR = 8.51, 95%CI: 1.78-40.54; P value = 0.007; AA vs GG; and HR = 5.61, 95%CI: 1.01-31.12; P value = 0.048; AA vs AG).
The same analysis was performed for AGT2 (rs4762) AA genotype and DAE development. A statistically significant increased risk in the probability of DAE was observed in the Northern Italy cohort (HR = 5.97, 95%CI: 1.32-27.01; P value = 0.02; AA vs GG) and when considering all but the Northern Italy cohort together as a unique cohort (HR = 3.12, 95%CI: 1.2-8.14; P value = 0.02; AA vs AG, and HR = 2.73, 95%CI: 1.18-6.32: P value = 0.02; AA vs GG).
No statistically significant effect on survival was found for AGT1 (rs699) or AGT2 (rs4762) using univariate or multivariate models in any cohort or combination thereof (Supplementary Table 6 and Supplementary Table 7).
The aim of Precision Oncology is to decide the treatment to be recommended to a specific patient according to the individualized evaluation of the clinical, biochemical and hopefully, molecular profile. It is common to focus all the attention on the genomic abnormalities of cancer to define the best intervention, but it is well known that, patients’ genetic background, irrespective of the tumor, is involved in the efficacy and safety of any therapeutic intervention. The best example is the clearance related to the glucuronidation activity resulting in fast and slow elimination of drugs and their metabolites[18]. Response to inflammation or tolerance to antiangiogenic agents is also influenced by genetic background and most cancer treatments have targets affecting several of these separate domains. In some instances, these non-cancer effects may become a surrogate of drug activity and even be correlated with improved outcomes as already described in the introduction.
This multicenter international study explored whether specific genetic variants, as identified by SNP analysis, may be linked to the development of AEs that have been associated with improved outcome. This is not only the case for DAEs in patients with HCC treated with sorafenib[12,19], as has been extensively proven, but also when using other TKIs such as regorafenib[20]. Furthermore, the association of DAEs with improved outcome is also being reported when using chemotherapy or immunotherapy not only in liver cancer but also in other tumor types[3-5].
The results of our multicenter study confirm that the genetic background of patients plays a key role in the emergence of specific events that are linked to a distinct outcome under HCC treatment. Previously, different SNPs were reported to be potentially associated with survival outcomes[16,17] while others were identified as significantly associated with a higher likelihood of DAEs affecting the angiotensin gene and its AGT2 (rs4762) variant.
Our results confirmed that the distribution of the AGT genetic variants studied, AGT1 (rs699) and AGT2 (rs4762), was comparable across patients from northern and southern Italy and those from Barcelona, and confirmed that the frequency of reference and alternative alleles follow the reported distribution for the European population[21,22].
Although rs699 and rs4762 could not be associated with AHT events in our patients, the most relevant finding is the identification of AGT2 (rs4762) AA genotype as a predictor of DAE development [HR = 5.97; P value = 0.0201] in the Northern Italy cohort and its validation in the remaining 3 cohorts when they were considered as one unique cohort [HR = 3.12 (95%CI: 1.2-8.14); P value = 0.02 and HR = 2.73 (95%CI: 1.18-6.32); P value = 0.02].
AGT2 (rs4762) is a missense variant that codes for the replacement of threonine by methionine with no reported clear association with blood AGT protein levels. AGT2 (rs4762) has been associated with renal dysplasia, a potentially benign disease[22]. However, published data suggest that rs4762 may be associated with an increased risk of mortality in patients with heart failure[23] and with the development of intracranial hemorrhage in stroke patients[24]. Available data at this moment do not allow to unequivocally associate an increase in blood AGT levels with rs4762 polymorphism, but it is speculated that it could induce Renin-Angiotensin System (RAS) activation. The RAS is a key regulator of systemic homeostasis by controlling salt-water balance and consequently, blood pressure. Interestingly, several studies have also unveiled the activation of this system in several peripheral tissues (tRAS)[25] and organs including skin and liver[26]. Since activation of tRAS is associated with tissue regeneration, inflammation and fibrosis[27] and considering that all of these are key components of tumor development, tRAS activation is likely to play a role in carcinogenesis. A review by Ager EI and collaborators[28] describes the potential contribution of tRAS activation in cancer development and progression putting the emphasis not only on tumor angiogenesis, but also on inflammation and fibrosis. Considering that the components of the tRAS pathway are also participating in physiological and pathological wound healing and fibrosis processes that are particularly important in skin homeostasis[29,30], DAE development in our patients with rs4762 AA genotype may be considered a consequence of tRAS activation at the skin level.
The role of genetic variants in the components of the RAS pathway has been extensively reported in the past years and some of these roles involve response to anti-neoplastic treatments, disease prognosis and patient survival. In that sense, it is already known that ACE I/D rs4646994, a variant of the Angiotensin-Converting Enzyme (ACE), has been associated with prediction of response to bevacizumab in metastatic breast and colorectal cancer patients[31]. The AGTrs5050 GG genotype[32] is reported to be linked to poor prognosis in patients with astrocytoma. A very interesting in silico study by Goswami and colleagues analyzed 354 SNPs in the AGT gene[33] in order to predict those variants that are pathogenic and how amino acid substitutions would impact protein function. In this study, AGT2 rs4762 was categorized mainly as a damaging AGT SNP with controversial results on its pathogenicity or disease identity. Thus, the importance of genetic variants is determined by the levels and/or functionality of the protein they code for. Along these lines, Feng et al[34] proposed that cancer tissue levels of ACE2 correlates with immune infiltrates and these would affect the prognosis of cancer patients. In another study, Urupet et al[35] suggested that low expression of the AGT gene and high expression of an HLA-class II gene (HLADQA1) were independent predictors associated with response in glioblastoma patients treated with bevacizumab.
AGT2 (rs4762) has been associated with an increased risk of AHT in several studies[36,37] but this association remains controversial as the results could not be confirmed in other series of individuals analyzed[38]. We were not able to identify an association between AGT2 rs4762 and AHT in our patients not even when analyzing the impact of concomitant medication that the BCLC1 and BCLC2 cohort patients received for AHT that included IEACA (renin angiotensin aldosterone axis inhibitor)(Supplementary Table 8). This could be related to the low frequency of AGT2 rs4762 in patients who developed this AE [0 (0%) in the BCLC1 and Northern Italy cohorts, 1 (1.27%) in the BCLC2 cohort and 2 (2.9%) in the Naples cohort].
However, in our cohort, the impact of AGT2 (rs4762) was maintained when the multivariate was adjusted for history of AHT.
To the best of our knowledge, the relationship between AGT2 rs4762AA genotype and DAE development in HCC patients under sorafenib treatment has not been previously reported. This is a ‘proof-of-concept’ study to identify a novel genetic marker to screen for patients with good outcome. It would be interesting for our results to be validated in other cancer types besides HCC or even in different therapeutic approaches. If this were to be the case, AGT2 (rs4762) should be considered a good prognosis marker instead of being only a predictor of DAE development. The retrospective profile of the study did not allow us to assess analysis related to radiological response as the radiological follow-up between the cohorts was different, and this could be seen as a limitation of the study. However, we prefer to be conservative and avoid overestimating the role of DAEs on the radiological outcome.
In conclusion, our findings open the window to explore individual genetic susceptibility as prognostic factors or predictors of treatment outcome, and to unveil novel mechanisms triggered by oncological treatment and their potential link to tumor response and patient survival.
DAE development in HCC patients receiving TKIs could be explained by the AGT2 (rs4762) gene variant. If validated in other anti-oncogenic treatments, it might be considered a good prognosis marker.
In hepatocellular carcinoma (HCC), patients regardless of the chosen treatment, the development of dermatologic adverse events (DAEs) is associated with better outcome. The underlying mechanism of these effects is unknown.
Distinct genetic variants could have an effect to the likelihood of developing DAEs in patients treated with TKIs for advanced HCC.
The objective of this study was to evaluate the association of two specific AGT gene single-nucleotide polymorphisms, rs699 and rs4762, in DAE development.
Four cohorts were used to assess the effect, as training and external validation, of the effect of AGT1 (rs699) and AGT2 (rs4762) on the development of DAEs in patients with advanced HCC.
AGT2 (rs4762) AA genotype was related to an increased risk of DAEs development in the Northern Italy cohort in a multivariate model adjusted for clinically relevant factors such as BCLC stage, ECOG-PS, diabetes and arterial hypertension (AHT). This effect was externally validated in the validation cohort (combining BCLC1, BCLC2 and Naples cohorts).
The development of DAEs in patients treated with TKIs for advanced HCC could be explained by the AGT2 (rs4762) SNP.
The AGT2 (rs4762) SNP could be proposed as a valuable predictive marker if a similar effect is found in other anti-oncogenic treatments.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng H, China; Sahin TT, Turkey A-Editor: Vasudevan A S-Editor: Ma YJ L-Editor: Webster JR P-Editor: Ma YJ
| 1. | Gordon KB, Tajuddin A, Guitart J, Kuzel TM, Eramo LR, VonRoenn J. Hand-foot syndrome associated with liposome-encapsulated doxorubicin therapy. Cancer. 1995;75:2169-2173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Nagore E, Insa A, Sanmartín O. Antineoplastic therapy-induced palmar plantar erythrodysesthesia ('hand-foot') syndrome. Incidence, recognition and management. Am J Clin Dermatol. 2000;1:225-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 197] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 3. | LB Saltz, M Kies, JL Abbruzzesse, N Azarnia, M Needle, L Saltz JA. The presence and intensity of the cetuximab-induced acne-like rash predicts increased survival in studies across multiple malignancies. Proc Am Soc Clin Oncol 2003; 22: 204. |
| 4. | Naidoo J, Page DB, Li BT, Connell LC, Schindler K, Lacouture ME, Postow MA, Wolchok JD. Toxicities of the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann Oncol. 2015;26:2375-2391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 978] [Cited by in RCA: 1056] [Article Influence: 105.6] [Reference Citation Analysis (0)] |
| 5. | Freeman-Keller M, Kim Y, Cronin H, Richards A, Gibney G, Weber JS. Nivolumab in Resected and Unresectable Metastatic Melanoma: Characteristics of Immune-Related Adverse Events and Association with Outcomes. Clin Cancer Res. 2016;22:886-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 661] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 6. | Li Y, Gao ZH, Qu XJ. The adverse effects of sorafenib in patients with advanced cancers. Basic Clin Pharmacol Toxicol. 2015;116:216-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 7. | Lai SE, Kuzel T, Lacouture ME. Hand-foot and stump syndrome to sorafenib. J Clin Oncol. 2007;25:341-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | Jacobi U, Waibler E, Schulze P, Sehouli J, Oskay-Ozcelik G, Schmook T, Sterry W, Lademann J. Release of doxorubicin in sweat: first step to induce the palmar-plantar erythrodysesthesia syndrome? Ann Oncol. 2005;16:1210-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Robert C, Soria JC, Spatz A, Le Cesne A, Malka D, Pautier P, Wechsler J, Lhomme C, Escudier B, Boige V, Armand JP, Le Chevalier T. Cutaneous side-effects of kinase inhibitors and blocking antibodies. Lancet Oncol. 2005;6:491-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 370] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 10. | Berner F, Bomze D, Diem S, Ali OH, Fässler M, Ring S, Niederer R, Ackermann CJ, Baumgaertner P, Pikor N, Cruz CG, van de Veen W, Akdis M, Nikolaev S, Läubli H, Zippelius A, Hartmann F, Cheng HW, Hönger G, Recher M, Goldman J, Cozzio A, Früh M, Neefjes J, Driessen C, Ludewig B, Hegazy AN, Jochum W, Speiser DE, Flatz L. Association of Checkpoint Inhibitor-Induced Toxic Effects With Shared Cancer and Tissue Antigens in Non-Small Cell Lung Cancer. JAMA Oncol. 2019;5:1043-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 295] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 11. | Ruiz-Pinto S, Pita G, Martín M, Nuñez-Torres R, Cuadrado A, Shahbazi MN, Caronia D, Kojic A, Moreno LT, de la Torre-Montero JC, Lozano M, López-Fernández LA, Ribelles N, García-Saenz JA, Alba E, Milne RL, Losada A, Pérez-Moreno M, Benítez J, González-Neira A. Regulatory CDH4 Genetic Variants Associate With Risk to Develop Capecitabine-Induced Hand-Foot Syndrome. Clin Pharmacol Ther. 2021;109:462-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Reig M, Torres F, Rodriguez-Lope C, Forner A, LLarch N, Rimola J, Darnell A, Ríos J, Ayuso C, Bruix J. Early dermatologic adverse events predict better outcome in HCC patients treated with sorafenib. J Hepatol. 2014;61:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 13. | Rimola J, Díaz-González Á, Darnell A, Varela M, Pons F, Hernandez-Guerra M, Delgado M, Castroagudin J, Matilla A, Sangro B, Rodriguez de Lope C, Sala M, Gonzalez C, Huertas C, Minguez B, Ayuso C, Bruix J, Reig M. Complete response under sorafenib in patients with hepatocellular carcinoma: Relationship with dermatologic adverse events. Hepatology. 2018;67:612-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 14. | Corominas J, Sapena V, Sanduzzi-Zamparelli M, Millán C, Samper E, Llarch N, Iserte G, Torres F, Da Fonseca LG, Muñoz-Martínez S, Forner A, Bruix J, Boix L, Reig M. Activated Lymphocytes and Increased Risk of Dermatologic Adverse Events during Sorafenib Therapy for Hepatocellular Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | European Association for the Study of The Liver. European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4501] [Article Influence: 346.2] [Reference Citation Analysis (2)] |
| 16. | Casadei Gardini A, Marisi G, Faloppi L, Scarpi E, Foschi FG, Iavarone M, Lauletta G, Corbelli J, Valgiusti M, Facchetti F, Della Corte C, Neri LM, Tamberi S, Cascinu S, Scartozzi M, Amadori D, Nanni O, Tenti E, Ulivi P, Frassineti GL. eNOS polymorphisms and clinical outcome in advanced HCC patients receiving sorafenib: final results of the ePHAS study. Oncotarget. 2016;7:27988-27999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 17. | Marisi G, Petracci E, Raimondi F, Faloppi L, Foschi FG, Lauletta G, Iavarone M, Canale M, Valgiusti M, Neri LM, Ulivi P, Orsi G, Rovesti G, Vukotic R, Conti F, Cucchetti A, Ercolani G, Andrikou K, Cascinu S, Scartozzi M, Casadei-Gardini A. ANGPT2 and NOS3 Polymorphisms and Clinical Outcome in Advanced Hepatocellular Carcinoma Patients Receiving Sorafenib. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Wilkinson GR. Drug Metabolism and Variability among Patients in Drug Response. 2005. |
| 19. | Díaz-González Á, Sanduzzi-Zamparelli M, Sapena V, Torres F, LLarch N, Iserte G, Forner A, da Fonseca L, Ríos J, Bruix J, Reig M. Systematic review with meta-analysis: the critical role of dermatological events in patients with hepatocellular carcinoma treated with sorafenib. Aliment Pharmacol Ther. 2019;49:482-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Bruix J, Merle P, Granito A, Huang Y-H, Bodoky G, Yokosuka O, Rosmorduc O, Breder VV, Gerolami R, Masi G, Ross PJ, Qin S, Song T, Bronowicki J-P, Ollivier-Hourmand I, Kudo M, Xu L, Baumhauer A, Meinhardt G, Han G. Hand-foot skin reaction (HFSR) and overall survival (OS) in the phase 3 RESORCE trial of regorafenib for treatment of hepatocellular carcinoma (HCC) progressing on sorafenib. J Clin Oncol. 2018;36:412-412. [RCA] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. |
rs699 RefSNP Report-dbSNP-NCBI [Internet].
[cited 2021 Mar 3]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs699#frequency_tab, |
| 22. | rs4762 RefSNP Report-dbSNP-NCBI [Internet]. [cited 2021 Mar 3]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs4762#clinical_significance. |
| 23. | Pilbrow AP, Palmer BR, Frampton CM, Yandle TG, Troughton RW, Campbell E, Skelton L, Lainchbury JG, Richards AM, Cameron VA. Angiotensinogen M235T and T174M gene polymorphisms in combination doubles the risk of mortality in heart failure. Hypertension. 2007;49:322-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Park HK, Kim MC, Kim SM, Jo DJ. Assessment of two missense polymorphisms (rs4762 and rs699) of the angiotensinogen gene and stroke. Exp Ther Med. 2013;5:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Bader M. Tissue renin-angiotensin-aldosterone systems: Targets for pharmacological therapy. Annu Rev Pharmacol Toxicol. 2010;50:439-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 242] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 26. | Hunyady L, Catt KJ. Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Mol Endocrinol. 2006;20:953-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 402] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 27. | Deshayes F, Nahmias C. Angiotensin receptors: a new role in cancer? Trends Endocrinol Metab. 2005;16:293-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 318] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 28. | Ager EI, Neo J, Christophi C. The renin-angiotensin system and malignancy. Carcinogenesis. 2008;29:1675-1684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 228] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 29. | Bernasconi R, Nyström A. Balance and circumstance: The renin angiotensin system in wound healing and fibrosis. Cell Signal. 2018;51:34-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Steckelings UM, Wollschläger T, Peters J, Henz BM, Hermes B, Artuc M. Human skin: source of and target organ for angiotensin II. Exp Dermatol. 2004;13:148-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Moreno-Muñoz D, de la Haba-Rodríguez JR, Conde F, López-Sánchez LM, Valverde A, Hernández V, Martínez A, Villar C, Gómez-España A, Porras I, Rodríguez-Ariza A, Aranda E. Genetic variants in the renin-angiotensin system predict response to bevacizumab in cancer patients. Eur J Clin Invest. 2015;45:1325-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Perdomo-Pantoja A, Mejía-Pérez SI, Reynoso-Noverón N, Gómez-Flores-Ramos L, Soto-Reyes E, Sánchez-Correa TE, Guerra-Calderas L, Castro-Hernandez C, Vidal-Millán S, Sánchez-Corona J, Taja-Chayeb L, Gutiérrez O, Cacho-Diaz B, Alvarez-Gomez RM, Gómez-Amador JL, Ostrosky-Wegman P, Corona T, Herrera-Montalvo LA, Wegman-Ostrosky T. Angiotensinogen rs5050 germline genetic variant as potential biomarker of poor prognosis in astrocytoma. PLoS One. 2018;13:e0206590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Goswami AM. Computational analyses prioritize and reveal the deleterious nsSNPs in human angiotensinogen gene. Comput Biol Chem. 2020;84:107199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Feng H, Wei X, Pang L, Wu Y, Hu B, Ruan Y, Liu Z, Liu J, Wang T. Prognostic and Immunological Value of Angiotensin-Converting Enzyme 2 in Pan-Cancer. Front Mol Biosci. 2020;7:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Urup T, Michaelsen SR, Olsen LR, Toft A, Christensen IJ, Grunnet K, Winther O, Broholm H, Kosteljanetz M, Issazadeh-Navikas S, Poulsen HS, Lassen U. Angiotensinogen and HLA class II predict bevacizumab response in recurrent glioblastoma patients. Mol Oncol. 2016;10:1160-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 36. | Jeunemaitre X, Gimenez-Roqueplo AP, Célérier J, Corvol P. Angiotensinogen variants and human hypertension. Curr Hypertens Rep. 1999;1:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | International Consortium for Blood Pressure Genome-Wide Association Studies; Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjögren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimäki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND; CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium, Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kähönen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Köttgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grässler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stančáková A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikäinen LP, Soininen P, Tukiainen T, Würtz P, Ong RT, Dörr M, Kroemer HK, Völker U, Völzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Järvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1554] [Cited by in RCA: 1560] [Article Influence: 111.4] [Reference Citation Analysis (0)] |
| 38. | Charita B, Padma G, Sushma P, Deepak P, Padma T. Estimation of risk and interaction of single nucleotide polymorphisms at angiotensinogen locus causing susceptibility to essential hypertension: a case control study. J Renin Angiotensin Aldosterone Syst. 2012;13:461-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |